Accelerate computational Bayesian inference workflows in Rthrough interactive modelling, visualization, and inference. The packageleverages directed acyclic graphs (DAGs) to create a unified languagelanguage for business stakeholders, statisticians, and programmers. Dueto its visual nature and simple model construction,causactserves as a great entry-point for newcomers to computational Bayesianinference.
The
causactpackage offers robust support for bothfoundational and advanced Bayesian models. While introductory models arewell-covered, the utilization of multi-variate distributions such asmulti-variate normal, multi-nomial, or dirichlet distributions, may notwork as expected. There are ongoing enhancements in the pipeline tofacilitate construction of these more intricate models.

While proficiency in R is the only requirement for users of thispackage, it also functions as a introductory probabilistic programminglanguage, seamlessly incorporating thenumpyro Pythonpackage to facilitate Bayesian inference without the need to learn anysyntax outside of the package or R. Furthermore, the package streamlinesthe process of indexing categorical variables, which often presents acomplex syntax hurdle for those new to computational Bayesianmethods.
For enhanced learning, thecausact package for Bayesianinference is featured inA Business Analyst's Introduction to Business Analyticsavailable athttps://www.causact.com/.
Feedback and encouragement is appreciated via github issues.
To install thecausact package, follow the stepsoutlined below:
For the current stable release, which is tailored to integrate withPython’snumpyro package, employ the following command:
install.packages("causact")Then, seeEssentialDependencies if you want to be able to automate sampling using thenumpyro package.
If you want the most recent development version (not recommended),execute the following:
install.packages("remotes")remotes::install_github("flyaflya/causact")To harness the full potential ofcausact for DAGvisualization and Bayesian posterior analysis, it’s vital to ensureproper integration with thenumpyro package. Given thePython-based nature ofnumpyro, a few essentialdependencies must be in place. Execute the following commands afterinstallingcausact:
library(causact)install_causact_deps()If prompted, respond withY to any inquiries related toinstalling miniconda.
Note: If opting for installation on Posit Cloud,temporarily adjust your project’s RAM to 4GB during the installationprocess (remember to APPLY CHANGES). This preemptive measure helps avoidencountering an
Error: Error creating conda environment [exit code 137].After installation, feel free to revert the settings to 1GB of RAM.
Note: The September 11, 2023 release of
reticulate(v1.32) has caused an issue whichgives aTypeError: the first argument must be callableerror when usingdag_numpyro()on windows. If youexperience this, install the dev version ofreticulatebyfollowing the below steps:
Install RTOOLS by using installer at:https://cran.r-project.org/bin/windows/Rtools/
Run this to get the dev version of
reticulate:
# install DEV version of reticulate# install.packages("pak") #uncomment as neededpak::pak("rstudio/reticulate")In cases where legacy compatibility is paramount and you still relyon the operationality of thedag_greta() function, considerinstallingv0.4.2 of thecausact package.However, it’s essential to emphasize that this approach isnotrecommended for general usage:
### EXERCISECAUTION BEFORE EXECUTING THESE LINES### Only proceed if dag_greta() is integral to your existing codebaseinstall.packages("remotes")remotes::install_github("flyaflya/causact@v0.4.2")Your judicious choice of installation method will ensure a seamlessand effective integration of thecausact package into yourcomputational toolkit.
Example taken fromhttps://www.causact.com/graphical-models-tell-joint-distribution-stories.html#graphical-models-tell-joint-distribution-storieswith the packagesdag_foo() functions further describedhere:
#> Initializing python, numpyro, and other dependencies. This may take up to 15 seconds...#> Initializing python, numpyro, and other dependencies. This may take up to 15 seconds...COMPLETED!#> #> Attaching package: 'causact'#> The following objects are masked from 'package:stats':#> #> binomial, poisson#> The following objects are masked from 'package:base':#> #> beta, gammalibrary(causact)graph=dag_create()%>%dag_node(descr ="Get Card",label ="y",rhs =bernoulli(theta),data = carModelDF$getCard)%>%dag_node(descr ="Card Probability",label ="theta",rhs =beta(2,2),child ="y")%>%dag_plate(descr ="Car Model",label ="x",data = carModelDF$carModel,nodeLabels ="theta",addDataNode =TRUE)graph%>%dag_render()
graph%>%dag_render(shortLabel =TRUE)
numpyro codedrawsDF= graph%>%dag_numpyro()drawsDF### see top of data frame#> # A tibble: 4,000 × 4#> theta_Toyota.Corolla theta_Subaru.Outback theta_Kia.Forte theta_Jeep.Wrangler#> <dbl> <dbl> <dbl> <dbl>#> 1 0.215 0.582 0.226 0.839#> 2 0.194 0.643 0.264 0.853#> 3 0.216 0.602 0.243 0.831#> 4 0.217 0.647 0.177 0.843#> 5 0.186 0.580 0.309 0.853#> 6 0.230 0.633 0.237 0.846#> 7 0.217 0.619 0.263 0.859#> 8 0.205 0.692 0.274 0.862#> 9 0.191 0.693 0.332 0.847#> 10 0.188 0.655 0.174 0.851#> # ℹ 3,990 more rowsNote: if you have used older versions of causact,please know that dag_numpyro() is a drop-in replacement fordag_greta().
drawsDF%>%dagp_plot()
Credible interval plots.
numpyro code without executing it (for debugging orlearning)numpyroCode= graph%>%dag_numpyro(mcmc =FALSE)#>#> ## The below code will return a posterior distribution#> ## for the given DAG. Use dag_numpyro(mcmc=TRUE) to return a#> ## data frame of the posterior distribution:#> reticulate::py_run_string("#> import numpy as np#> import numpyro as npo#> import numpyro.distributions as dist#> import pandas as pd#> from jax import random#> from numpyro.infer import MCMC, NUTS#> from jax.numpy import transpose as t#> from jax.numpy import (exp, log, log1p, expm1, abs, mean,#> sqrt, sign, round, concatenate, atleast_1d,#> cos, sin, tan, cosh, sinh, tanh,#> sum, prod, min, max, cumsum, cumprod )#> ## note that above is from JAX numpy package, not numpy.#>#> y = np.array(r.carModelDF.getCard) #DATA#> x = pd.factorize(np.array(r.carModelDF.carModel),use_na_sentinel=True)[0] #DIM#> x_dim = len(np.unique(x)) #DIM#> x_crd = pd.factorize(np.array(r.carModelDF.carModel),use_na_sentinel=True)[1] #DIM#> def graph_model(y,x):#> ## Define random variables and their relationships#> with npo.plate('x_dim',x_dim):#> theta = npo.sample('theta', dist.Beta(2,2))#>#> y = npo.sample('y', dist.Bernoulli(theta[x]),obs=y)#>#>#> # computationally get posterior#> mcmc = MCMC(NUTS(graph_model), num_warmup = 1000, num_samples = 4000)#> rng_key = random.PRNGKey(seed = 1234567)#> mcmc.run(rng_key,y,x)#> axis_labels = {'x_dim': x_crd}#> rv_to_axes = {'theta': ['x_dim']}#> drop_names = set()#> samples = mcmc.get_samples(group_by_chain=True) # dict: name -> [chains, draws, *axes]#> import numpy as np, pandas as pd, string#> # Flatten (chains*draws) and expand RVs using rv_to_axes + axis_labels#> flat = {name: np.reshape(val, (-1,) + val.shape[2:]) for name, val in list(samples.items())}#> out = {}#> allowed = set(string.ascii_letters + string.digits + '._')#> def sanitize_label(s):#> s = str(s).strip().replace(' ', '.') # spaces -> dots#> s = ''.join((ch if ch in allowed else '.') for ch in s)#> # collapse repeated dots without regex#> while '..' in s:#> s = s.replace('..', '.')#> s = s.strip('.')#> return s if s else '1'#>#> for name, arr in list(flat.items()):#> # Drop deterministic RVs entirely#> if name in drop_names:#> continue#> axes = rv_to_axes.get(name, []) # [] if not present#> # If scalar per draw, keep as a single column#> if arr.ndim == 1:#> out[name] = arr#> continue#> # Otherwise (vector/matrix/...): expand to separate columns#> trailing = arr.shape[1:]#> arr2 = arr.reshape(arr.shape[0], int(np.prod(trailing)))#> for j in range(arr2.shape[1]):#> idx = np.unravel_index(j, trailing)#> parts = []#> for axis, i in enumerate(idx):#> if axis < len(axes):#> axis_name = axes[axis]#> labels = axis_labels.get(axis_name)#> if labels is not None:#> labs = np.asarray(labels).astype(str)#> lab = labs[i] if i < labs.shape[0] else str(i + 1)#> parts.append(sanitize_label(lab))#> else:#> parts.append(str(i + 1))#> else:#> parts.append(str(i + 1))#> # No brackets; join labels with '_' so tibble won't append trailing dots#> col = name if not parts else name + '_' + '_'.join(parts)#> out[col] = arr2[:, j]#> drawsDF = pd.DataFrame(out)"#> ) ##END PYTHON STRING#> drawsDF = reticulate::py$drawsDFWhether you encounter a clear bug, have a suggestion for improvement,or just have a question, we are thrilled to help you out. In all cases,please file aGitHub issue. Ifreporting a bug, please include a minimal reproducible example.
We welcome help turningcausact into the most intuitiveand fastest method of converting stakeholder narratives aboutdata-generating processes into actionable insight from posteriordistributions. If you want to help us achieve this vision, we welcomeyour contributions after reading thenewcontributor guide. Please note that this project is released with aContributorCode of Conduct. By participating in this project you agree to abideby its terms.
For more info, seeA Business Analyst's Introduction to Business Analyticsavailable athttps://www.causact.com. You can also check out thepackage’s vignette:vignette("narrative-to-insight-with-causact"). Twoadditional examples are shown below.
McElreath, Richard. Statistical rethinking: A Bayesian course withexamples in R and Stan. Chapman and Hall/CRC, 2018.
library(tidyverse)library(causact)# data object used below, chimpanzeesDF, is built-in to causact packagegraph=dag_create()%>%dag_node("Pull Left Handle","L",rhs =bernoulli(p),data = causact::chimpanzeesDF$pulled_left)%>%dag_node("Probability of Pull","p",rhs =1/ (1+exp(-((alpha+ gamma+ beta)))),child ="L")%>%dag_node("Actor Intercept","alpha",rhs =normal(alphaBar, sigma_alpha),child ="p")%>%dag_node("Block Intercept","gamma",rhs =normal(0,sigma_gamma),child ="p")%>%dag_node("Treatment Intercept","beta",rhs =normal(0,0.5),child ="p")%>%dag_node("Actor Population Intercept","alphaBar",rhs =normal(0,1.5),child ="alpha")%>%dag_node("Actor Variation","sigma_alpha",rhs =exponential(1),child ="alpha")%>%dag_node("Block Variation","sigma_gamma",rhs =exponential(1),child ="gamma")%>%dag_plate("Observation","i",nodeLabels =c("L","p"))%>%dag_plate("Actor","act",nodeLabels =c("alpha"),data = chimpanzeesDF$actor,addDataNode =TRUE)%>%dag_plate("Block","blk",nodeLabels =c("gamma"),data = chimpanzeesDF$block,addDataNode =TRUE)%>%dag_plate("Treatment","trtmt",nodeLabels =c("beta"),data = chimpanzeesDF$treatment,addDataNode =TRUE)graph%>%dag_render(width =2000,height =800)
graph%>%dag_render(shortLabel =TRUE)
drawsDF= graph%>%dag_numpyro()drawsDF%>%dagp_plot()
Gelman, Andrew, Hal S. Stern, John B. Carlin, David B. Dunson, AkiVehtari, and Donald B. Rubin. Bayesian data analysis. Chapman andHall/CRC, 2013.
library(tidyverse)library(causact)# data object used below, schoolDF, is built-in to causact packagegraph=dag_create()%>%dag_node("Treatment Effect","y",rhs =normal(theta, sigma),data = causact::schoolsDF$y)%>%dag_node("Std Error of Effect Estimates","sigma",data = causact::schoolsDF$sigma,child ="y")%>%dag_node("Exp. Treatment Effect","theta",child ="y",rhs = avgEffect+ schoolEffect)%>%dag_node("Pop Treatment Effect","avgEffect",child ="theta",rhs =normal(0,30))%>%dag_node("School Level Effects","schoolEffect",rhs =normal(0,30),child ="theta")%>%dag_plate("Observation","i",nodeLabels =c("sigma","y","theta"))%>%dag_plate("School Name","school",nodeLabels ="schoolEffect",data = causact::schoolsDF$schoolName,addDataNode =TRUE)graph%>%dag_render()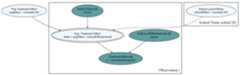
drawsDF= graph%>%dag_numpyro()drawsDF%>%dagp_plot()