

You can install {gtreg} with the following code.
install.packages("gtreg")You can install the development version of {gtreg} fromGitHub with:
# install.packages("devtools")devtools::install_github("shannonpileggi/gtreg")The {gtreg} package creates tabular data summaries appropriate forregulatory submissions. The package builds the tables using{gtsummary}.
Here areslidesand arecorded talk (17 min)from “Introducing {gtreg}: an R package to produce regulatory tables forclinical research” presented at the 2022 R in Medicine conference.
Summarize Raw Adverse Counts
tbl_ae_count() provides counts of all AEs, and omitspercentage statistics as multiple AEs can occur per subject.
library(gtreg)tbl_ae_count<- df_adverse_events|>tbl_ae_count(ae = adverse_event,soc = system_organ_class,by = drug_attribution )|>add_overall(across ="by")|>modify_spanning_header(all_ae_cols()~"**Drug Attribution**")|>bold_labels()
Summarize Adverse Events by Grade
tbl_ae() counts one AE per subject by maximum grade;percentage statistics are provided by default with the denominatorsreflecting the number of patients in the study.
library(gtreg)gtsummary::theme_gtsummary_compact()#> Setting theme "Compact"tbl_ae<- df_adverse_events|>tbl_ae(id_df = df_patient_characteristics,id = patient_id,ae = adverse_event,soc = system_organ_class,by = grade,strata = trt )|>modify_header(all_ae_cols()~"**Grade {by}**")|>bold_labels()
Focus on rates of high grade complications
tbl_ae_focus() also counts one AE per subject by maximumgrade, and is a convenience to summarize dichotomous AE attributes.
tbl_ae_focus<- df_adverse_events|>tbl_ae_focus(id_df = df_patient_characteristics,id = patient_id,ae = adverse_event,include =c(any_complication, grade3_complication) )
Regulatory summary
tbl_reg_summary() creates a data summary table oftenseen in regulatory submissions.
tbl_reg_summary<- df_patient_characteristics|>tbl_reg_summary(by = trt,include =c(marker, status))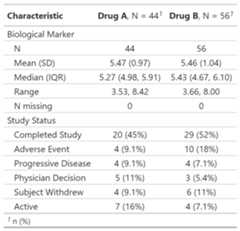
Print an AE listing
tbl_listing() creates a gtsummary-class listing of datato enable grouped printing.
tbl_listing<-head(df_adverse_events,n =10)|>select(system_organ_class, adverse_event, grade, drug_attribution, patient_id)|> dplyr::arrange(adverse_event,desc(grade))|>tbl_listing(group_by = system_organ_class)|>bold_labels()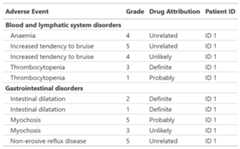
Please note that the gtreg project is released with aContributorCode of Conduct. By contributing to this project, you agree to abideby its terms.