A set of tools for Bayesian nonparametric density estimation usingMartingale posterior distributions including theCopulaResampling (CopRe) algorithm. Also included are a Gibbssampler for the marginal Gibbs-type mixture model and an extension toinclude full uncertainty quantification via a predictiveSequenceResampling (SeqRe) algorithm.The CopRe and SeqRe samplers generate random nonparametric distributionsas output, leading to complete nonparametric inference on posteriorsummaries. Routines for calculating arbitrary functionals from thesampled distributions are included as well as an important algorithm forfinding the number and location of modes, which can then be used toestimate the clusters in the data.
Installation
You can install the latest stable development version of CopRe fromGitHub with:
# install.packages("devtools")devtools::install_github("blakemoya/copre")Usage
CopRe
The basic usage of CopRe for density estimation is to supply a datavector, a number of forward simulations per sample, and a number ofsamples to draw:
library(copre)data<-sample(c(rnorm(100,mean =-2),rnorm(100,mean =2)))res_cop<-copre(data,250,100)plot(res_cop)+geom_function(fun =function(x) (dnorm(x,mean =-2)+dnorm(x,mean =2))/2 )
Currently only a Gaussian kernel copula is supported but more optionsare to be added in future versions.
SeqRe
Using SeqRe first involves specifying a model via a base measureG and an exchangeable sequence measureSq.Here we can set up a normal location scale mixture with componentsassigned according to a Dirichlet process.
We can then fit the marginal mixture model with thegibbsmix MCMC routine and then extend the results to theirfull nonparametric potential viaseqre.
b_norm<-G_normls()s_dp<-Sq_dirichlet()res_seq<-gibbsmix(data,100, b_norm, s_dp)|>seqre()plot(res_seq)+geom_function(fun =function(x) (dnorm(x,mean =-2)+dnorm(x,mean =2))/2 )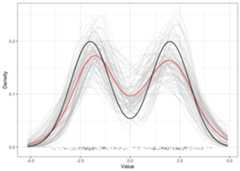
Utilities
The moments of the estimated distributions can be obtained by callingmoment, and arbitrary functionals of interest can beobtained similarly withfunctionals.
moms<-data.frame(Mean =moment(res_cop,1),Variance =moment(res_cop,2))with(moms,qplot(Mean, Variance)+theme_bw() )
The functionmodes can be used to isolate local maximaand minima in the density estimates. Here we can see the detected modesin black and the antimodes in red. Experiment with using sampleantimodes as a distribution over cluster boundaries!
res_cop.dens<-grideval(res_cop)ns<-modes(res_cop.dens,idx =TRUE)us<-antimodes(res_cop.dens,idx =TRUE)p<-plot(res_cop.dens)for (iin1:length(res_cop)) { p<- p+geom_point(data =data.frame(x = ns[[i]]$value,y = res_cop.dens[i, ns[[i]]$idx]),aes(x = x,y = y),shape =24,size =0.5,alpha =0.5)+geom_point(data =data.frame(x = us[[i]]$value,y = res_cop.dens[i, us[[i]]$idx]),aes(x = x,y = y),shape =25,size =0.5,alpha =0.5,color ='red')}print(p)
Feedback
If you have encounter any bugs or other problems while using CopRe,let me know using the Issues tab. For new feature requests, contact mevia email atblakemoya@utexas.edu.
[8]ページ先頭