The goal of ecan is to support ecological analysis.
install.packages("ecan")# development# install.packages("devtools")devtools::install_github("matutosi/ecan")You can use almost the same functionality in shiny.
https://matutosi.shinyapps.io/ecanvis/ .
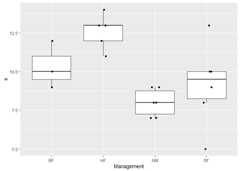
library(ecan)library(vegan)#> Loading required package: permute#> Loading required package: lattice#> This is vegan 2.6-4library(dplyr)#>#> Attaching package: 'dplyr'#> The following objects are masked from 'package:stats':#>#> filter, lag#> The following objects are masked from 'package:base':#>#> intersect, setdiff, setequal, unionlibrary(stringr)library(tibble)library(ggplot2)data(dune)data(dune.env)df<-table2df(dune)%>% dplyr::left_join(tibble::rownames_to_column(dune.env,"stand"))#> Joining with `by = join_by(stand)`sp_dammy<- tibble::tibble("species"=colnames(dune),"dammy_1"= stringr::str_sub(colnames(dune),1,1),"dammy_6"= stringr::str_sub(colnames(dune),6,6))df<- df%>% dplyr::left_join(sp_dammy)#> Joining with `by = join_by(species)`df#> # A tibble: 197 × 10#> stand species abundance A1 Moisture Manage…¹ Use Manure dammy_1 dammy_6#> <chr> <chr> <dbl> <dbl> <ord> <fct> <ord> <ord> <chr> <chr>#> 1 1 Achimill 1 2.8 1 SF Hayp… 4 A i#> 2 1 Elymrepe 4 2.8 1 SF Hayp… 4 E e#> 3 1 Lolipere 7 2.8 1 SF Hayp… 4 L e#> 4 1 Poaprat 4 2.8 1 SF Hayp… 4 P a#> 5 1 Poatriv 2 2.8 1 SF Hayp… 4 P i#> 6 2 Achimill 3 3.5 1 BF Hayp… 2 A i#> 7 2 Alopgeni 2 3.5 1 BF Hayp… 2 A e#> 8 2 Bellpere 3 3.5 1 BF Hayp… 2 B e#> 9 2 Bromhord 4 3.5 1 BF Hayp… 2 B o#> 10 2 Elymrepe 4 3.5 1 BF Hayp… 2 E e#> # … with 187 more rows, and abbreviated variable name ¹Managementdiv<-shdi(df)%>% dplyr::left_join(select_one2multi(df,"stand"))#> Joining with `by = join_by(stand)`group<-"Management"div_index<-"s"div%>%ggplot(aes(x = .data[[group]],y = .data[[div_index]]))+geom_boxplot(outlier.shape =NA)+# do not show outer pointgeom_jitter(height =0,width =0.1)
ind_val(df,group ="Moisture",row_data =TRUE)#> $relfrq#> 1 2 3 4#> Achimill 0.7142857 0.50 0.0000000 0.0#> Elymrepe 0.4285714 0.50 0.0000000 0.5#> Lolipere 1.0000000 0.75 0.1428571 0.5#> Poaprat 1.0000000 1.00 0.2857143 0.5#> Poatriv 0.7142857 0.75 0.4285714 1.0#> Alopgeni 0.1428571 0.50 0.4285714 1.0#> Bellpere 0.4285714 0.75 0.0000000 0.0#> Bromhord 0.4285714 0.50 0.0000000 0.0#> Scorautu 0.8571429 1.00 0.8571429 1.0#> Trifrepe 0.8571429 0.75 0.7142857 1.0#> Agrostol 0.0000000 0.50 0.8571429 1.0#> Bracruta 0.7142857 0.75 0.7142857 1.0#> Cirsarve 0.0000000 0.25 0.0000000 0.0#> Sagiproc 0.1428571 0.25 0.4285714 1.0#> Anthodor 0.4285714 0.50 0.1428571 0.0#> Planlanc 0.7142857 0.50 0.0000000 0.0#> Rumeacet 0.4285714 0.00 0.0000000 1.0#> Trifprat 0.4285714 0.00 0.0000000 0.0#> Juncbufo 0.1428571 0.00 0.1428571 1.0#> Eleopalu 0.0000000 0.00 0.7142857 0.0#> Juncarti 0.0000000 0.00 0.5714286 0.5#> Ranuflam 0.0000000 0.00 0.8571429 0.0#> Vicilath 0.2857143 0.25 0.0000000 0.0#> Hyporadi 0.1428571 0.25 0.1428571 0.0#> Chenalbu 0.0000000 0.00 0.1428571 0.0#> Comapalu 0.0000000 0.00 0.2857143 0.0#> Callcusp 0.0000000 0.00 0.4285714 0.0#> Airaprae 0.0000000 0.25 0.1428571 0.0#> Salirepe 0.1428571 0.00 0.2857143 0.0#> Empenigr 0.0000000 0.00 0.1428571 0.0#>#> $relabu#> 1 2 3 4#> Achimill 0.48780488 0.5121951 0.00000000 0.00000000#> Elymrepe 0.25531915 0.2978723 0.00000000 0.44680851#> Lolipere 0.46204620 0.3927393 0.05280528 0.09240924#> Poaprat 0.35036496 0.3576642 0.08759124 0.20437956#> Poatriv 0.24806202 0.2713178 0.15503876 0.32558140#> Alopgeni 0.02846975 0.2241993 0.19928826 0.54804270#> Bellpere 0.40000000 0.6000000 0.00000000 0.00000000#> Bromhord 0.39506173 0.6049383 0.00000000 0.00000000#> Scorautu 0.33922261 0.2226148 0.24028269 0.19787986#> Trifrepe 0.27636364 0.2290909 0.18909091 0.30545455#> Agrostol 0.00000000 0.2818792 0.38926174 0.32885906#> Bracruta 0.29197080 0.1532847 0.24817518 0.30656934#> Cirsarve 0.00000000 1.0000000 0.00000000 0.00000000#> Sagiproc 0.05161290 0.2258065 0.18064516 0.54193548#> Anthodor 0.33333333 0.5185185 0.14814815 0.00000000#> Planlanc 0.70588235 0.2941176 0.00000000 0.00000000#> Rumeacet 0.50000000 0.0000000 0.00000000 0.50000000#> Trifprat 1.00000000 0.0000000 0.00000000 0.00000000#> Juncbufo 0.06060606 0.0000000 0.09090909 0.84848485#> Eleopalu 0.00000000 0.0000000 1.00000000 0.00000000#> Juncarti 0.00000000 0.0000000 0.50000000 0.50000000#> Ranuflam 0.00000000 0.0000000 1.00000000 0.00000000#> Vicilath 0.63157895 0.3684211 0.00000000 0.00000000#> Hyporadi 0.19047619 0.3333333 0.47619048 0.00000000#> Chenalbu 0.00000000 0.0000000 1.00000000 0.00000000#> Comapalu 0.00000000 0.0000000 1.00000000 0.00000000#> Callcusp 0.00000000 0.0000000 1.00000000 0.00000000#> Airaprae 0.00000000 0.5384615 0.46153846 0.00000000#> Salirepe 0.27272727 0.0000000 0.72727273 0.00000000#> Empenigr 0.00000000 0.0000000 1.00000000 0.00000000#>#> $indval#> 1 2 3 4#> Achimill 0.348432056 0.25609756 0.000000000 0.00000000#> Elymrepe 0.109422492 0.14893617 0.000000000 0.22340426#> Lolipere 0.462046205 0.29455446 0.007543612 0.04620462#> Poaprat 0.350364964 0.35766423 0.025026069 0.10218978#> Poatriv 0.177187154 0.20348837 0.066445183 0.32558140#> Alopgeni 0.004067107 0.11209964 0.085409253 0.54804270#> Bellpere 0.171428571 0.45000000 0.000000000 0.00000000#> Bromhord 0.169312169 0.30246914 0.000000000 0.00000000#> Scorautu 0.290762241 0.22261484 0.205956588 0.19787986#> Trifrepe 0.236883117 0.17181818 0.135064935 0.30545455#> Agrostol 0.000000000 0.14093960 0.333652924 0.32885906#> Bracruta 0.208550574 0.11496350 0.177267987 0.30656934#> Cirsarve 0.000000000 0.25000000 0.000000000 0.00000000#> Sagiproc 0.007373272 0.05645161 0.077419355 0.54193548#> Anthodor 0.142857143 0.25925926 0.021164021 0.00000000#> Planlanc 0.504201681 0.14705882 0.000000000 0.00000000#> Rumeacet 0.214285714 0.00000000 0.000000000 0.50000000#> Trifprat 0.428571429 0.00000000 0.000000000 0.00000000#> Juncbufo 0.008658009 0.00000000 0.012987013 0.84848485#> Eleopalu 0.000000000 0.00000000 0.714285714 0.00000000#> Juncarti 0.000000000 0.00000000 0.285714286 0.25000000#> Ranuflam 0.000000000 0.00000000 0.857142857 0.00000000#> Vicilath 0.180451128 0.09210526 0.000000000 0.00000000#> Hyporadi 0.027210884 0.08333333 0.068027211 0.00000000#> Chenalbu 0.000000000 0.00000000 0.142857143 0.00000000#> Comapalu 0.000000000 0.00000000 0.285714286 0.00000000#> Callcusp 0.000000000 0.00000000 0.428571429 0.00000000#> Airaprae 0.000000000 0.13461538 0.065934066 0.00000000#> Salirepe 0.038961039 0.00000000 0.207792208 0.00000000#> Empenigr 0.000000000 0.00000000 0.142857143 0.00000000#>#> $maxcls#> Achimill Elymrepe Lolipere Poaprat Poatriv Alopgeni Bellpere Bromhord#> 1 4 1 2 4 4 2 2#> Scorautu Trifrepe Agrostol Bracruta Cirsarve Sagiproc Anthodor Planlanc#> 1 4 3 4 2 4 2 1#> Rumeacet Trifprat Juncbufo Eleopalu Juncarti Ranuflam Vicilath Hyporadi#> 4 1 4 3 3 3 1 2#> Chenalbu Comapalu Callcusp Airaprae Salirepe Empenigr#> 3 3 3 2 3 3#>#> $indcls#> Achimill Elymrepe Lolipere Poaprat Poatriv Alopgeni Bellpere#> 0.34843206 0.22340426 0.46204620 0.35766423 0.32558140 0.54804270 0.45000000#> Bromhord Scorautu Trifrepe Agrostol Bracruta Cirsarve Sagiproc#> 0.30246914 0.29076224 0.30545455 0.33365292 0.30656934 0.25000000 0.54193548#> Anthodor Planlanc Rumeacet Trifprat Juncbufo Eleopalu Juncarti#> 0.25925926 0.50420168 0.50000000 0.42857143 0.84848485 0.71428571 0.28571429#> Ranuflam Vicilath Hyporadi Chenalbu Comapalu Callcusp Airaprae#> 0.85714286 0.18045113 0.08333333 0.14285714 0.28571429 0.42857143 0.13461538#> Salirepe Empenigr#> 0.20779221 0.14285714#>#> $pval#> Achimill Elymrepe Lolipere Poaprat Poatriv Alopgeni Bellpere Bromhord#> 0.240 0.417 0.062 0.378 0.502 0.053 0.131 0.190#> Scorautu Trifrepe Agrostol Bracruta Cirsarve Sagiproc Anthodor Planlanc#> 0.797 0.698 0.383 0.614 0.286 0.072 0.311 0.107#> Rumeacet Trifprat Juncbufo Eleopalu Juncarti Ranuflam Vicilath Hyporadi#> 0.092 0.136 0.004 0.024 0.214 0.001 0.699 1.000#> Chenalbu Comapalu Callcusp Airaprae Salirepe Empenigr#> 1.000 0.449 0.072 0.755 0.594 1.000#>#> $error#> [1] 0#>#> attr(,"class")#> [1] "indval"ind_val(df,group ="Management")#> Joining with `by = join_by(numeric_Management)`#> # A tibble: 30 × 4#> Management species ind.val p.value#> <fct> <chr> <dbl> <dbl>#> 1 SF Elymrepe 0.188 0.684#> 2 SF Alopgeni 0.547 0.038#> 3 SF Agrostol 0.472 0.054#> 4 SF Cirsarve 0.167 1#> 5 SF Sagiproc 0.241 0.514#> 6 SF Chenalbu 0.167 1#> 7 BF Achimill 0.386 0.118#> 8 BF Lolipere 0.45 0.07#> 9 BF Poaprat 0.379 0.188#> 10 BF Bellpere 0.362 0.126#> # … with 20 more rowsind_val(df,group ="Use")#> Joining with `by = join_by(numeric_Use)`#> # A tibble: 30 × 4#> Use species ind.val p.value#> <ord> <chr> <dbl> <dbl>#> 1 Haypastu Elymrepe 0.292 0.288#> 2 Haypastu Lolipere 0.259 0.796#> 3 Haypastu Poaprat 0.288 0.824#> 4 Haypastu Poatriv 0.451 0.118#> 5 Haypastu Alopgeni 0.359 0.184#> 6 Haypastu Agrostol 0.269 0.589#> 7 Haypastu Cirsarve 0.125 1#> 8 Haypastu Sagiproc 0.178 0.8#> 9 Haypastu Juncbufo 0.118 0.848#> 10 Haypastu Chenalbu 0.125 1#> # … with 20 more rowsind_val(df,group ="Manure")#> Joining with `by = join_by(numeric_Manure)`#> # A tibble: 30 × 4#> Manure species ind.val p.value#> <ord> <chr> <dbl> <dbl>#> 1 4 Elymrepe 0.5 0.048#> 2 4 Lolipere 0.351 0.211#> 3 4 Poaprat 0.315 0.28#> 4 4 Bellpere 0.248 0.469#> 5 4 Cirsarve 0.333 0.279#> 6 2 Achimill 0.309 0.262#> 7 2 Poatriv 0.299 0.394#> 8 2 Bromhord 0.173 0.703#> 9 2 Anthodor 0.178 0.763#> 10 2 Rumeacet 0.522 0.041#> # … with 20 more rowslibrary(ggdendro)library(dendextend)#> Registered S3 method overwritten by 'dendextend':#> method from#> rev.hclust vegan#>#> ---------------------#> Welcome to dendextend version 1.16.0#> Type citation('dendextend') for how to cite the package.#>#> Type browseVignettes(package = 'dendextend') for the package vignette.#> The github page is: https://github.com/talgalili/dendextend/#>#> Suggestions and bug-reports can be submitted at: https://github.com/talgalili/dendextend/issues#> You may ask questions at stackoverflow, use the r and dendextend tags:#> https://stackoverflow.com/questions/tagged/dendextend#>#> To suppress this message use: suppressPackageStartupMessages(library(dendextend))#> ---------------------#>#> Attaching package: 'dendextend'#> The following object is masked from 'package:ggdendro':#>#> theme_dendro#> The following object is masked from 'package:permute':#>#> shuffle#> The following object is masked from 'package:stats':#>#> cutreecls<-cluster(dune,c_method ="average",d_method ="euclidean")ggdendro::ggdendrogram(cls)
indiv<-"stand"group<-"Use"ggdendro::ggdendrogram(cls_add_group(cls, df, indiv, group))#> Joining with `by = join_by(stand)`
col<-cls_color(cls, df, indiv, group)#> Joining with `by = join_by(stand)`#> Joining with `by = join_by(Use)`cls<-cls_add_group(cls, df, indiv, group)%>% stats::as.dendrogram()#> Joining with `by = join_by(stand)`labels_colors(cls)<-gray(0)plot(cls)dendextend::colored_bars(colors = col, cls, group,y_shift =0,y_scale =3)par(new =TRUE)plot(cls)
ord_dca<-ordination(dune,o_method ="dca")ord_pca<- df%>%df2table()%>%ordination(o_method ="pca")ord_dca_st<-ord_extract_score(ord_dca,score ="st_scores")ord_dca_st%>%ggplot(aes(DCA1, DCA2,label =rownames(.)))+geom_text()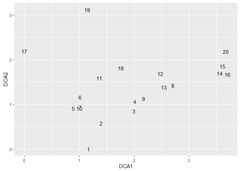
indiv<-"species"group<-"dammy_1"ord_pca_sp<-ord_add_group(ord_pca,score ="sp_scores", df, indiv, group)#> Joining with `by = join_by(species)`ord_pca_sp%>%ggplot(aes(PC1, PC2,label =rownames(.)))+geom_point(aes(col = .data[[group]]),alpha =0.4,size =7)+geom_text()+theme_bw()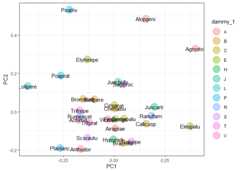
Toshikazu Matsumura (2022) Ecological analysis tools with R.https://github.com/matutosi/ecan/.