TheMFO package have been designed to calculate theMaximal Fat Oxidation (MFO), the exercise intensity that elicits MFO(Fatmax) and the SIN model to represent the fat oxidation kinetics.Three variables can be obtained from the SIN model: dilatation (d),symmetry (s) and traslation (t).Additionally, the package allows tocalculate MFO and Fatmax of multiple subjects.
This is a basic example which shows you how to use the MFOpackage:
library(devtools)install_github("JorgeDelro/MFO")library(MFO)First, we have to load the data which consists in 3 databases: -basal_df: basal metabolism database. - MFO_df: MFO test database. -VO2max_df: results of a graded exercise test of which the VO2max of thesubject is going to be extracted.
# Read dfsdata(list =c("basal_df","MFO_df","VO2max_df"),package ="MFO")# Convert to data.framebasal_df<-data.frame(basal_df)MFO_df<-data.frame(MFO_df)VO2max_df<-data.frame(VO2max_df)Then, we can used the function MFO
result_MFO<-MFO(step_time =20,db_MFO = MFO_df,db_basal = basal_df,db_graded = VO2max_df,cv_var ="RER",author ="Frayn",VO2max =NULL)and the MFO can be plotted
print(result_MFO$MFO_plot)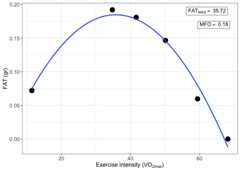
MFO kinetics are calculated using a database returned from MFOfunction called MFO_db.
result_MFO_kinetics<-MFO_kinetics(result_MFO$MFO_db)And again the function returns a plot with the results calculated
print(result_MFO_kinetics$MFO_kinetics_plot)