

You can install the package from CRAN:
install.packages("edibble")You can install the development version with:
# install.packages("remotes")remotes::install_github("emitanaka/edibble")The goal ofedibble R-package is to aid in the plan,design and simulation of experiments by mapping fundamental componentsof experiments to an object oriented system. Theedibblesystem is built on the principle that the system must make it easy torecover experimental context by encouraging the user to be explicitabout experimental details in fundamental terms.
Consider an experiment where you want to know what is an effectiveway of teaching (flipped or traditional style) for teaching a particularsubject and how different forms of exams (take-home, open-book orclosed-book) affect student’s marks.
There are four classes for this subject with each class holding 30students. The teaching style can only be applied to the whole class butexam can be different for individual students.
library(edibble)set.seed(2020)des<-design(name ="Effective teaching")%>%set_units(class =4,student =nested_in(class,30))%>%set_trts(style =c("flipped","traditional"),exam =c("take-home","open-book","closed-book"))%>%allot_trts(style~ class, exam~ student)%>%assign_trts("random")serve_table(des)#> # An edibble: 120 x 4#> class student style exam#> <U(4)> <U(120)> <T(2)> <T(3)>#> <chr> <chr> <chr> <chr>#> 1 class1 student001 traditional take-home#> 2 class1 student002 traditional take-home#> 3 class1 student003 traditional take-home#> 4 class1 student004 traditional open-book#> 5 class1 student005 traditional closed-book#> 6 class1 student006 traditional closed-book#> 7 class1 student007 traditional closed-book#> 8 class1 student008 traditional open-book#> 9 class1 student009 traditional open-book#> 10 class1 student010 traditional open-book#> # ℹ 110 more rowsBefore constructing the experiment, you might want to think aboutwhat you are recording for which level of unit and what values thesevariables can be recorded as.
out<- des%>%set_rcrds_of(student =c("exam_mark","quiz1_mark","quiz2_mark","gender"),class =c("room","teacher"))%>%expect_rcrds(exam_mark<=100, exam_mark>=0, quiz1_mark<= 15L, quiz1_mark>= 0L, quiz2_mark<= 30L, quiz2_mark>= 0L,factor(gender,levels =c("female","male","non-binary","unknown")))%>%serve_table()out#> # An edibble: 120 x 10#> class student style exam exam_mark quiz1_mark quiz2_mark gender#> <U(4)> <U(120)> <T(2)> <T(3)> <R(120)> <R(120)> <R(120)> <R(120)>#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>#> 1 class1 student001 traditio… take-ho… o o o o#> 2 class1 student002 traditio… take-ho… o o o o#> 3 class1 student003 traditio… take-ho… o o o o#> 4 class1 student004 traditio… open-bo… o o o o#> 5 class1 student005 traditio… closed-… o o o o#> 6 class1 student006 traditio… closed-… o o o o#> 7 class1 student007 traditio… closed-… o o o o#> 8 class1 student008 traditio… open-bo… o o o o#> 9 class1 student009 traditio… open-bo… o o o o#> 10 class1 student010 traditio… open-bo… o o o o#> # ℹ 110 more rows#> # ℹ 2 more variables: room <R(4)>, teacher <R(4)>When you export the above edibble design using theexport_design function, the variables you are recording areconstraint to the values you expect, e.g. for factors, the cells have adrop-down menu to select from possible values.
export_design(out,file ="/PATH/TO/FILE.xlsx")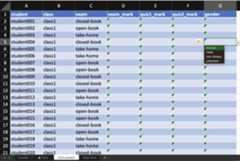
In addition, there is a spreadsheet for every observational level.E.g. hereroom andteacher is the same for allstudents in one class so rather than entering duplicate information,these are exported to another sheet for data entry.

There is also support for more complex nesting structures. You canalways make the structure using edibble and take the resulting dataframe to use in other experimental design software. It’s also possibleto bring existing data frame into edibble if you want to take advantageof the exporting feature in edibble.
design("nesting structure")%>%# there are 3 sites labelled A, B, Cset_units(site =c("A","B","C"),# each site has 2 blocks except B with 3 sitesblock =nested_in(site,"B"~3, .~2),# levels can be specified by their number instead# so for below "block1" has 30 plots,# "block2" and "block3" has 40 plots,# the rest of blocks have 20 plots.plot =nested_in(block,1~30,c(2,3)~40, .~20))%>%serve_table()#> # nesting structure#> # An edibble: 190 x 3#> site block plot#> <U(3)> <U(7)> <U(190)>#> <chr> <chr> <chr>#> 1 A block1 plot001#> 2 A block1 plot002#> 3 A block1 plot003#> 4 A block1 plot004#> 5 A block1 plot005#> 6 A block1 plot006#> 7 A block1 plot007#> 8 A block1 plot008#> 9 A block1 plot009#> 10 A block1 plot010#> # ℹ 180 more rowstidyverse is well suited for the data science project workflow asillustrated below in (B) (fromGrolemund and Wickham2017). For experimental data, the statistical aspect begins beforeobtaining data as depicted below in (A). The focus ofedibble is to facilitate work in (A).

The edibble R-package differ considerably to other packages forconstructing experimental design with a focus on the whole process andless on the randomisation process (which the other software generallyfocus and do well on). Some features include:
Currently, edibble:
The way that edibble specifies experimental design is largely novel(if I say so myself) and there are no work that resembles it. I’mconcurrently working on two extension packages:
deggust - to visualise the designs constructed fromedibble as ggplot2 objects (WIP).sizzled - for experiments that require sample sizecalculation (WIP).Below are some other related work. You can also have a look at theCRAN Task Viewfor Design of Experiment and Analysis of Experimental Data for awhole collection.
DeclareDesign for survey or sampling designsdesignr for balanced factorial designs with crossed andnested random and fixed effect to data framedae for functions useful in the design and ANOVA ofexperiments (this is in fact powering the randomisation in edibble)plotdesignr for designing agronomic fieldexperimentsedibble is hugely inspired by the work ofTidyverseTeam. I’m grateful for the dedication and work by the TidyverseTeam, as well asRDevelopment Core Team that supports the core R ecosystem, that madedeveloping this package possible.
The implementation in edibble adopt a similar nomenclature and designphilosophy as tidyverse (and where it does not, it’s likely myshortcoming) so that tidyverse users can leverage their familiarity ofthe tidyverse language when using edibble. Specifically, edibble followsthe philosophy:
<verb>_<noun> (e.g. set_units andset_rcrds) where the nouns are generally plural. Exceptionsare when the subject matter is clearly singular(e.g. design andset_context);tibble foradditions to edibble graph;dplyr::case_when (LHS is character or integer for edibblehowever).Please note that the edibble project is released with aContributorCode of Conduct. By contributing to this project, you agree to abideby its terms.